In the post-genomic era, we are faced with the daunting challenge of translating all of this genomic information into cures for human diseases. One of the major bottlenecks for drug discovery is that much of the genome remains uncharacterized, hampering our efforts to uncover their biological or therapeutic functions. Another challenge is that many of the protein targets that we know to be drivers of human diseases belong to a category known as “undruggable,” meaning that we have no means of developing pharmacological or biologic tools against the target for developing a therapeutic.
Activity-based protein profiling (ABPP) is an exciting technology that is enabling researchers to identify new therapeutic targets and even develop new and safer pharmacological agents for therapeutic discovery. ABPP uses active-site directed chemical probes to directly assess the functional states of large numbers of proteins directly in complex biological systems. These activity-based probes consist of a chemical warhead that binds to functional sites within proteins and a handle which can be used for visualization of these targets by fluorescence or identification of these targets by mass spectrometry-based proteomics. When applied to various disease settings, ABPP has been successfully used to discover many promising therapeutic targets for diseases such as cancer, inflammation, depression and anxiety, obesity, and neurodegenerative diseases. An additional unique feature of this ABPP technology is that it also enables the development of inhibitors against protein targets that are targeted by activity-based probes, even those that are uncharacterized and considered to be undruggable. Because the activity-based probes are binding to functional sites within proteins, small-molecule inhibitors can be competed against the probe binding, enabling inhibitor discovery and accelerating the process of drug discovery. In addition, because the probes themselves are binding across many proteins in-parallel, the selectivity of these inhibitors can be assessed on a proteome-wide level. Developing selective inhibitors that specifically inhibit the therapeutic target but not unrelated off-targets help in mitigating toxicities and side-effects, thus contributing to new and safer drugs.
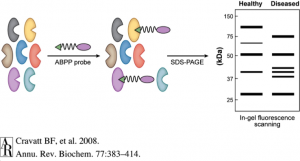
ABPP, started in Benjamin Cravatt’s laboratory at The Scripps Research Institute, has contributed to several interesting discoveries. For example, Cravatt and Daniel Nomura, now an Associate Professor at UC Berkeley, demonstrated that the enzyme monoacylglycerol lipase (MAGL) controls a fatty acid network that contributes to tumor growth (6). Their study showed that MAGL could be a target for cancer treatment and, curiously, suggested that there is a link between a high-fat diet and cancer progression. Nomura and Cravatt also showed that MAGL inhibition in the brain leads to elevations in endogenous cannabinoid lipid levels that act on the cannabinoid receptors (the same receptor that THC from marijuana binds) and also lowers pro-inflammatory eicosanoids lipid levels to inhibit inflammation and protect and neurodegeneration in the brain. Very potent and selective MAGL inhibitors have been developed using the ABPP technology and have recently entered Phase I clinical trials in humans through Abide Therapeutics in La Jolla, CA. Nomura’s lab at Berkeley has continued to identify novel therapeutic targets and potential therapeutic leads for treating cancer using the ABPP technology.
Hopefully, in the near future, drugs created thanks to ABPP will be successful in clinical trials.
Acknowledgments
Daniel Nomura has critically read and edited the blog post, sharing his valuable insights into ABPP technology.
References
1. National Cancer Institute. SEER Stat Fact Sheets: All Cancer Sites. http://seer.cancer.gov/statfacts/html/all.html (accessed Nov 22, 2015).
2. The Scripps Research Institute. The Cravatt Lab Research. http://www.scripps.edu/cravatt/research.html (accessed Nov 22, 2015).
3. Medina-Cleghorn, D.; Nomura, D. K. Exploring Metabolic Pathways and Regulation through Functional Chemoproteomic and Metabolomic Platforms. Chemistry & Biology. 2014, 21, 1171-1184. http://www.sciencedirect.com/science/article/pii/S1074552114002361 (accessed Nov 22, 2015).
4. Cravatt, B. F.; Wright, A. T.; Kozarich, J. W. Activity-Based Protein Profiling: From Enzyme Chemistry to Proteomic Chemistry. Annu. Rev. Biochem. 2008, 77, 383-414. http://www.annualreviews.org/doi/pdf/10.1146/annurev.biochem.75.101304.124125 (accessed Nov 22, 2015).
5. Bogyo, M. Finding enzymes that are actively involved in cancer. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 2379-2380. http://www.pnas.org/content/107/6/2379.full (accessed Nov 22, 2015).
6. Nomura, D. K.; Long, J. Z.; Niessen, S.; Hoover, H. S.; Ng, S. W.; Cravatt, B. F. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell, 2010, 140, 49-61. http://www.sciencedirect.com/science/article/pii/S0092867409014391 (accessed Nov 22, 2015).
7. Long et al. Selective Blockade of 2-Arachidonoylglycerol Hydrolysis Produces Cannabinoid Behavioral Effects. Nat. Chem. Biology. 2009, 5, 37-44. http://www.nature.com/nchembio/journal/v5/n1/full/nchembio.129.html (accessed Nov 22, 2015).
8. Special Feature. Greatest Hits. Nat. Chem. Biology. 2015, 11, 364-367. http://www.nature.com/nchembio/journal/v11/n6/full/nchembio.1815.html (accessed Nov 22, 2015).