Finding Its Target: How CRISPR Cas9 “Reads” DNA
By Sinead de Cleir
Since its application for gene editing was discovered in 2012 by Dr. Jennifer Doudna and Dr. Emmanuel Charpentier, clustered regularly interspaced short palindromic repeats, or CRISPR for short, has upended the scientific world. Although the use and ethical implications of CRISPR are still being explored, some mechanisms of this bacterial immune system remain a mystery. CRISPR can take six hours to search a genome of over four million base pairs, but how does it complete its search?
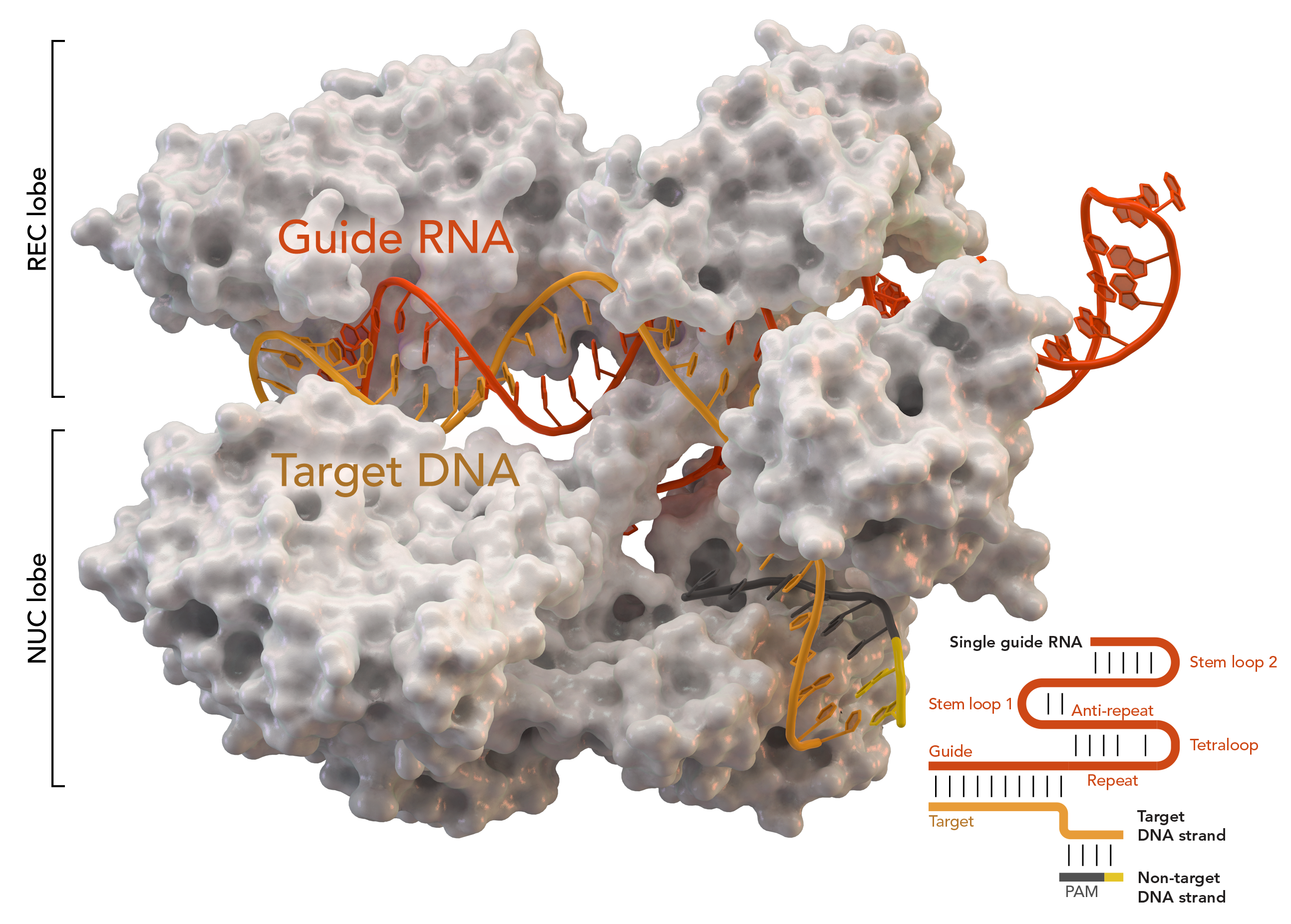
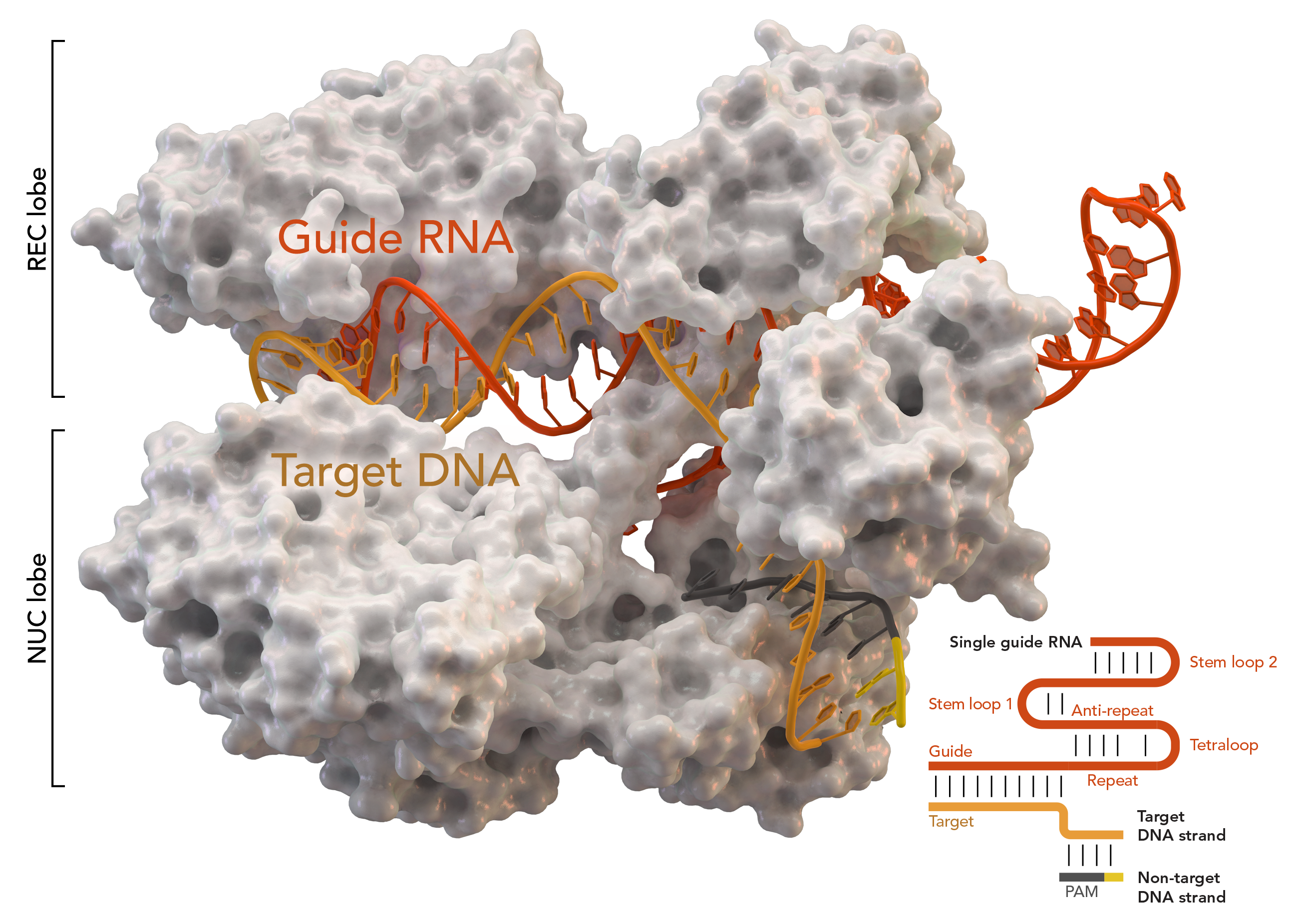
One key part of the CRISPR system is the CRISPR Cas9 (CRISPR-associated) protein. The Cas9 protein performs a target search during which it “reads” millions of DNA sequences, searching for the sequence that is both next to a protospacer-adjacent motif (PAM) and matches its single guide RNA (sgRNA). Once it finds the sequence, it initiates an “R-loop,” which is the base-pairing of the sgRNA to one DNA strand, at the PAM. Cas9 has an interrogation state—when it has engaged with the PAM but has yet to form the R-loop—that is key to its search for DNA. However, this state lasts about 30 milliseconds in bacteria, making the structure of the interrogation state thus far unknown.
To overcome the short duration of the interrogation complex, a team of researchers at UC Berkeley formed a disulfide bond between Cas9 and non-target DNA which did not match the sgRNA. Since the DNA was not the target, Cas9 would scan but not cleave. This bonded protein-DNA complex maintained the ability to cleave target DNA, proving that the bond did not disrupt Cas9’s ability. In order to visualize structures that are too small to image with light, the researchers used cryo-electron microscopy (EM) imaging to examine the complex as it did its search.
Cryo-EM imaging revealed two conformations of the complex: one with linear DNA, and one with bent DNA. The linear conformation represents a scanning state when Cas9 may just be beginning to engage with the PAM. After engaging with the PAM, Cas9 checks if the DNA sequence is complementary to its sgRNA. The bent conformation reveals the mechanism with which Cas9 “reads” DNA: it bends DNA. To achieve this desired conformation, Cas9’s lobes move to pinch DNA. Cas9, after finding the PAM, checks the possible complementary DNA by twisting it. This momentarily breaks the bonds between its base pairs and flips target-strand nucleotides toward the sgRNA so they can be checked for a match.
After discovering this mechanism, researchers needed to check if the bent conformation still appeared without the disulfide bond that stabilized the Cas9-DNA complex. They used interrogation complexes without the bond in a DNA cyclization assay. The assay results were consistent with a bend in the DNA, demonstrating that DNA is bent during Cas9’s search.
Having revealed the bent conformation of DNA, scientists worked on the further examination of DNA conformations through the comparison of the “native,” unmodified interrogation complex and the cross-linked complex. The phenomenon in which Cas9 breaks pairs to read them, known as protein-induced base unstacking, relates to an increase in permanganate reactivity. The induced native complex increases in permanganate reactivity at Thy(+1) and Thy(+2) whereas the induced cross-linked complex changes at Thy(+4) and Thy(+6). While both complexes bend DNA, the native complex distorts the nucleotides located right next to the PAM.
Understanding and learning more about how CRISPR Cas9 reads DNA is important for future applications of genome editing. Investigating the mechanism of CRISPR could improve its efficiency. Given that Cas9 reads DNA by twisting it, it is worthwhile to investigate whether existing bends in DNA could slow or quicken the genome search, depending on how they bend
References
Doudna, J.A., Cofsky, J.C., Soczek, K.M., Knott, G.J., & Nogales, E. (2021). CRISPR-Cas9 bends and twists DNA to read its sequence. bioRxiv.
Jones, D. L., Leroy, P., Unoson, C., Fange, D., Ćurić, V., Lawson, M. J., & Elf, J. (2017). Kinetics of dCas9 target search in Escherichia coli.
Science,
357(6358), 1420–1424.
https://doi.org/10.1126/science.aah7084
Image: Splettstoesser, T. (2016). Crispr-associated protein Cas9 (white) from Staphylococcus aureus based on Pdb Id. Wikimedia Commons. Retrieved March 10, 2022, from
https://commons.wikimedia.org/wiki/File:Cas9_5AXW.png